Building an efficient sparse keyword index in Python
Fast and accurate sparse keyword indexing
Semantic search is a new category of search built on recent advances in Natural Language Processing (NLP). Traditional search systems use keywords to find data. Semantic search has an understanding of natural language and identifies results that have the same meaning, not necessarily the same keywords.
While semantic search adds amazing capabilities, sparse keyword indexes can still add value. There may be cases where finding an exact match is important or we just want a fast index to quickly do an initial scan of a dataset.
Unfortunately, there aren't a ton of great options for a local Python-based keyword index library. Most of the options available don't scale and/or are highly inefficient, designed only for simple situations.
Given that Python is an interpreted language, it often gets a bad rap from a performance standpoint. In some cases, it's justified as Python can be memory hungry and has a global interpreter lock (GIL) that forces single thread execution. But it is possible to build performant Python on par with other languages.
This article will explore how to build an efficient sparse keyword index in Python and compare the results with other approaches.
Install dependencies
Install txtai and all dependencies.
# Install txtai
pip install txtai pytrec_eval rank-bm25 elasticsearch==7.10.1
Introducing the problem
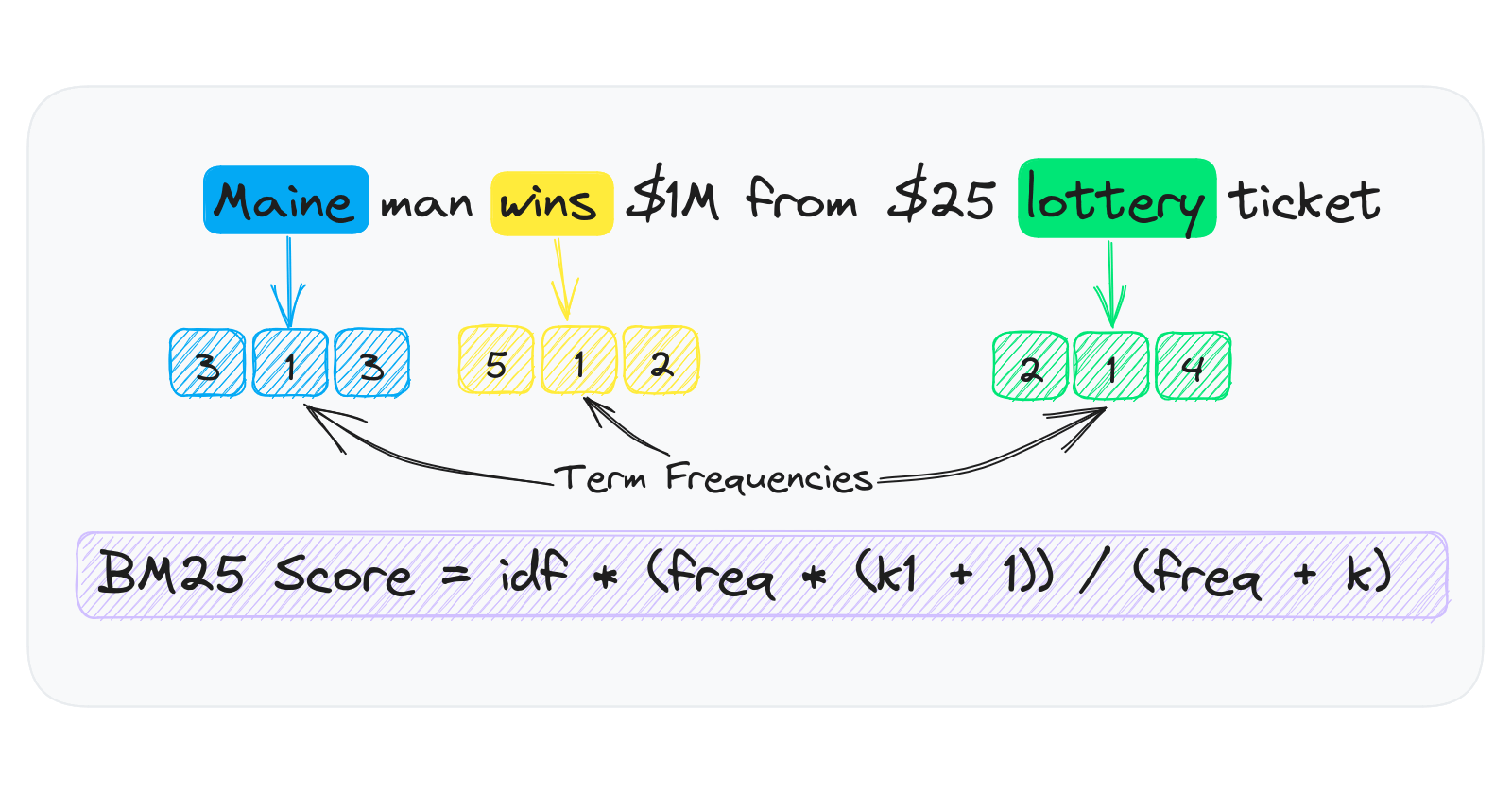
At a high level, keyword indexes work by tokenizing text into lists of tokens per document. These tokens are aggregated into frequencies per document and stored in term frequency sparse arrays.
The term frequency arrays are sparse given that they only store a frequency when the token exists in a document. For example, if a token exists in 1 of 1000 documents, the sparse array only has a single entry. A dense array stores 1000 entries all with zeros except for one.
One simple approach to store a term frequency sparse array in Python would be having a dictionary of {id: frequency} per token. The problem with this approach is that Python has significant object overhead.
Let's inspect the size used for a single number.
import sys
a = 100
sys.getsizeof(a)
28
28 bytes for a single integer. Compared to a native int/long which is 4 or 8 bytes, this is quite wasteful. Imagine having thousands of id: frequency mappings. Memory usage will grow fast.
Let's demonstrate. The code below runs a self contained Python process that creates a list of 10 million numbers.
Running as a separate process helps calculate more accurate memory usage stats.
import psutil
results = []
for x in range(int(1e7)):
results.append(x)
print(f"MEMORY USAGE = {psutil.Process().memory_info().rss / (1024 * 1024)} MB")
MEMORY USAGE = 394.640625 MB
Approximately 395 MB of memory is used for this array. That seems high.
Efficient numeric arrays in Python
Fortunately, Python has a module for building efficient arrays of numeric values. This module enables building arrays with the same native type.
Let's try doing that with a long long type, which takes 8 bytes.
from array import array
import psutil
results = array("q")
for x in range(int(1e7)):
results.append(x)
print(f"MEMORY USAGE = {psutil.Process().memory_info().rss / (1024 * 1024)} MB")
MEMORY USAGE = 88.54296875 MB
As we can see, memory usage went from 395 MB to 89 MB. That's a 4x reduction which is in line with the earlier calculate of 28 bytes/number vs 8 bytes/number.
Efficient processing of numeric data
Large computations in pure Python can also be painfully slow. Luckily, there is a robust landscape of options for numeric processing. The most popular framework is NumPy. There is also PyTorch and other GPU-based tensor processing frameworks.
Below is a simple example that sorts an array in Python vs NumPy to demonstrate.
import random
import time
data = [random.randint(1, 500) for x in range(1000000)]
start = time.time()
sorted(data, reverse=True)
print(time.time() - start)
0.33922290802001953
import numpy as np
data = np.array(data)
start = time.time()
np.sort(data)[::-1]
print(time.time() - start)
0.10296249389648438
As we can see, sorting an array in NumPy is significantly faster. It might not seem like a lot but this adds up when run in bulk.
Sparse keyword indexes in txtai
Now that we've discussed the key performance concepts, let's talk about how to apply this to building sparse keyword indexes.
Going back to the original approach for a term frequency sparse array, we see that using the Python array package is more efficient. In txtai, this method is used to build term frequency arrays for each token. This results in near native speed and memory usage.
The search method uses a number of NumPy methods to efficiently calculate query term matches. Each query is tokenized and those token term frequency arrays are retrieved to calculate query scores. These NumPy methods are all written in C and often drop the GIL. So once again, near native speed and the ability to use multithreading.
Read the full implementation on GitHub to learn more.
Evaluating performance
First, a review of the landscape. As said in the introduction, there aren't a ton of good options. Apache Lucene is by far the best traditional search index from a speed, performance and functionality standpoint. It's the base for Elasticsearch/OpenSearch and many other projects. But it requires Java.
Here are the options we'll explore.
Rank-BM25 project, the top result when searching for
python bm25.SQLite FTS5 extension. This extension builds a sparse keyword index right in SQLite.
We'll use the BEIR dataset. We'll also use a benchmarks script from the txtai project. This benchmarks script has methods to work with the BEIR dataset.
Couple important caveats on the benchmarks script.
For the SQLite FTS implementation, each token is joined together with an
ORclause. SQLite FTS implicitly joins clauses together withANDclauses by default. By contrast, Lucene's default operator is anOR.The Elasticsearch implementation uses 7.x as it's simpler to instantiate in a notebook.
All methods except Elasticsearch use txtai's unicode tokenizer to tokenize text for consistency
import os
# Get benchmarks script
os.system("wget https://raw.githubusercontent.com/neuml/txtai/master/examples/benchmarks.py")
# Create output directory
os.makedirs("beir", exist_ok=True)
# Download subset of BEIR datasets
datasets = ["trec-covid", "nfcorpus", "webis-touche2020", "scidocs", "scifact"]
for dataset in datasets:
url = f"https://public.ukp.informatik.tu-darmstadt.de/thakur/BEIR/datasets/{dataset}.zip"
os.system(f"wget {url}")
os.system(f"mv {dataset}.zip beir")
os.system(f"unzip -d beir beir/{dataset}.zip")
# Remove existing benchmark data
if os.path.exists("benchmarks.json"):
os.remove("benchmarks.json")
Now let's run the benchmarks.
# Remove existing benchmark data
if os.path.exists("benchmarks.json"):
os.remove("benchmarks.json")
# Runs benchmark evaluation
def evaluate(method):
for dataset in datasets:
command = f"python benchmarks.py beir {dataset} {method}"
print(command)
os.system(command)
# Calculate benchmarks
for method in ["bm25", "rank", "sqlite"]:
evaluate(method)
import json
import pandas as pd
def benchmarks():
# Read JSON lines data
with open("benchmarks.json") as f:
data = f.read()
df = pd.read_json(data, lines=True).sort_values(by=["source", "search"])
return df[["source", "method", "index", "memory", "search", "ndcg_cut_10", "map_cut_10", "recall_10", "P_10"]].reset_index(drop=True)
# Load benchmarks dataframe
df = benchmarks()
df[df.source == "trec-covid"].reset_index(drop=True)
| source | method | index | memory | search | ndcg_cut_10 | map_cut_10 | recall_10 | P_10 |
| trec-covid | bm25 | 101.96 | 997 | 0.28 | 0.58119 | 0.01247 | 0.01545 | 0.618 |
| trec-covid | sqlite | 60.16 | 880 | 23.09 | 0.56778 | 0.01190 | 0.01519 | 0.610 |
| trec-covid | rank | 61.75 | 3245 | 75.49 | 0.57773 | 0.01210 | 0.01550 | 0.632 |
df[df.source == "nfcorpus"].reset_index(drop=True)
| source | method | index | memory | search | ndcg_cut_10 | map_cut_10 | recall_10 | P_10 |
| nfcorpus | bm25 | 2.64 | 648 | 1.08 | 0.30639 | 0.11728 | 0.14891 | 0.21734 |
| nfcorpus | sqlite | 1.50 | 630 | 12.73 | 0.30695 | 0.11785 | 0.14871 | 0.21641 |
| nfcorpus | rank | 2.75 | 700 | 23.78 | 0.30692 | 0.11711 | 0.15320 | 0.21889 |
df[df.source == "webis-touche2020"].reset_index(drop=True)
| source | method | index | memory | search | ndcg_cut_10 | map_cut_10 | recall_10 | P_10 |
| webis-touche2020 | bm25 | 374.66 | 1137 | 0.37 | 0.36920 | 0.14588 | 0.22736 | 0.34694 |
| webis-touche2020 | sqlite | 220.46 | 1416 | 34.61 | 0.37194 | 0.14812 | 0.22890 | 0.35102 |
| webis-touche2020 | rank | 224.07 | 10347 | 81.22 | 0.39861 | 0.16492 | 0.23770 | 0.36122 |
df[df.source == "scidocs"].reset_index(drop=True)
| source | method | index | memory | search | ndcg_cut_10 | map_cut_10 | recall_10 | P_10 |
| scidocs | bm25 | 17.95 | 717 | 1.64 | 0.15063 | 0.08756 | 0.15637 | 0.0772 |
| scidocs | sqlite | 17.85 | 670 | 56.64 | 0.15156 | 0.08822 | 0.15717 | 0.0776 |
| scidocs | rank | 13.11 | 1056 | 162.99 | 0.14932 | 0.08670 | 0.15408 | 0.0761 |
df[df.source == "scifact"].reset_index(drop=True)
| source | method | index | memory | search | ndcg_cut_10 | map_cut_10 | recall_10 | P_10 |
| scifact | bm25 | 5.51 | 653 | 1.07 | 0.66324 | 0.61764 | 0.78761 | 0.087 |
| scifact | sqlite | 1.85 | 631 | 20.28 | 0.66630 | 0.61966 | 0.79494 | 0.088 |
| scifact | rank | 1.85 | 724 | 42.22 | 0.65618 | 0.61204 | 0.77400 | 0.085 |
The sections above show the metrics per source and method.
The table headers list the source (dataset), index method, index time(s), memory usage(MB), search time(s) and NDCG@10/MAP@10/RECALL@10/P@10 accuracy metrics. The tables are sorted by search time.
As we can see, txtai's implementation has the fastest search times across the board. But it is slower when it comes to index time. The accuracy metrics vary slightly but are all about the same per method.
Memory usage stands out. SQLite and txtai both have around the same usage per source. Rank-BM25 memory usage can get out of hand fast. For example, webis-touch2020, which is only ~400K records, uses 10 GB of memory compared to 700 MB for the other implementations.
Compare with Elasticsearch
Now that we've reviewed methods to build keyword indexes in Python, let's see how txtai's sparse keyword index compares to Elasticsearch.
We'll spin up an inline instance and run the same evaluations.
# Download and extract elasticsearch
os.system("wget https://artifacts.elastic.co/downloads/elasticsearch/elasticsearch-7.10.1-linux-x86_64.tar.gz")
os.system("tar -xzf elasticsearch-7.10.1-linux-x86_64.tar.gz")
os.system("chown -R daemon:daemon elasticsearch-7.10.1")
from subprocess import Popen, PIPE, STDOUT
# Start and wait for server
server = Popen(['elasticsearch-7.10.1/bin/elasticsearch'], stdout=PIPE, stderr=STDOUT, preexec_fn=lambda: os.setuid(1))
# Add benchmark evaluations for Elasticsearch
evaluate("es")
# Reload benchmarks dataframe
df = benchmarks()
df[df.source == "trec-covid"].reset_index(drop=True)
| source | method | index | memory | search | ndcg_cut_10 | map_cut_10 | recall_10 | P_10 |
| trec-covid | bm25 | 101.96 | 997 | 0.28 | 0.58119 | 0.01247 | 0.01545 | 0.618 |
| trec-covid | es | 71.24 | 636 | 2.09 | 0.59215 | 0.01261 | 0.01590 | 0.636 |
| trec-covid | sqlite | 60.16 | 880 | 23.09 | 0.56778 | 0.01190 | 0.01519 | 0.610 |
| trec-covid | rank | 61.75 | 3245 | 75.49 | 0.57773 | 0.01210 | 0.01550 | 0.632 |
df[df.source == "nfcorpus"].reset_index(drop=True)
| source | method | index | memory | search | ndcg_cut_10 | map_cut_10 | recall_10 | P_10 |
| nfcorpus | bm25 | 2.64 | 648 | 1.08 | 0.30639 | 0.11728 | 0.14891 | 0.21734 |
| nfcorpus | es | 3.95 | 627 | 11.47 | 0.30676 | 0.11761 | 0.14894 | 0.21610 |
| nfcorpus | sqlite | 1.50 | 630 | 12.73 | 0.30695 | 0.11785 | 0.14871 | 0.21641 |
| nfcorpus | rank | 2.75 | 700 | 23.78 | 0.30692 | 0.11711 | 0.15320 | 0.21889 |
df[df.source == "webis-touche2020"].reset_index(drop=True)
| source | method | index | memory | search | ndcg_cut_10 | map_cut_10 | recall_10 | P_10 |
| webis-touche2020 | bm25 | 374.66 | 1137 | 0.37 | 0.36920 | 0.14588 | 0.22736 | 0.34694 |
| webis-touche2020 | es | 168.28 | 629 | 0.62 | 0.37519 | 0.14819 | 0.22889 | 0.35102 |
| webis-touche2020 | sqlite | 220.46 | 1416 | 34.61 | 0.37194 | 0.14812 | 0.22890 | 0.35102 |
| webis-touche2020 | rank | 224.07 | 10347 | 81.22 | 0.39861 | 0.16492 | 0.23770 | 0.36122 |
df[df.source == "scidocs"].reset_index(drop=True)
| source | method | index | memory | search | ndcg_cut_10 | map_cut_10 | recall_10 | P_10 |
| scidocs | bm25 | 17.95 | 717 | 1.64 | 0.15063 | 0.08756 | 0.15637 | 0.0772 |
| scidocs | es | 11.07 | 632 | 10.25 | 0.14924 | 0.08671 | 0.15497 | 0.0765 |
| scidocs | sqlite | 17.85 | 670 | 56.64 | 0.15156 | 0.08822 | 0.15717 | 0.0776 |
| scidocs | rank | 13.11 | 1056 | 162.99 | 0.14932 | 0.08670 | 0.15408 | 0.0761 |
df[df.source == "scifact"].reset_index(drop=True)
| source | method | index | memory | search | ndcg_cut_10 | map_cut_10 | recall_10 | P_10 |
| scifact | bm25 | 5.51 | 653 | 1.07 | 0.66324 | 0.61764 | 0.78761 | 0.08700 |
| scifact | es | 2.90 | 625 | 9.62 | 0.66058 | 0.61518 | 0.78428 | 0.08667 |
| scifact | sqlite | 1.85 | 631 | 20.28 | 0.66630 | 0.61966 | 0.79494 | 0.08800 |
| scifact | rank | 1.85 | 724 | 42.22 | 0.65618 | 0.61204 | 0.77400 | 0.08500 |
Once again txtai's implementation compares well with Elasticsearch. The accuracy metrics vary but are all about the same.
It's important to note that in internal testing with solid state storage, Elasticsearch and txtai's speed is about the same. These times for Elasticsearch being a little slower are a product of running in a Google Colab environment.
Wrapping up
This notebook showed how to build an efficient sparse keyword index in Python. The benchmarks show that txtai provides a strong implementation both from an accuracy and speed standpoint, on par with Apache Lucene.
This keyword index can be used as a standalone index in Python or in combination with dense vector indexes to form a hybrid index.